Featured
Totally Wired: Where Could We Go With a Map of the Brain?
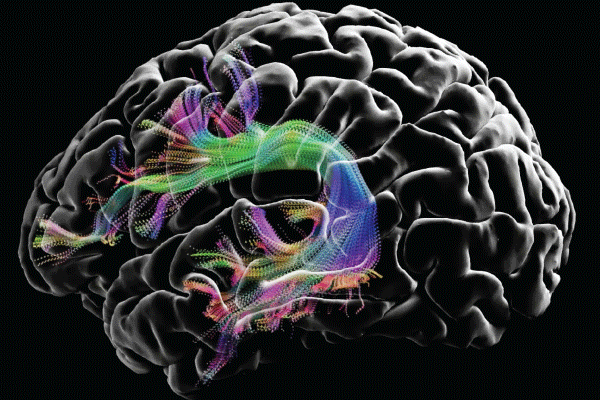
Mapping the brain's unknown territories and vibrant connections
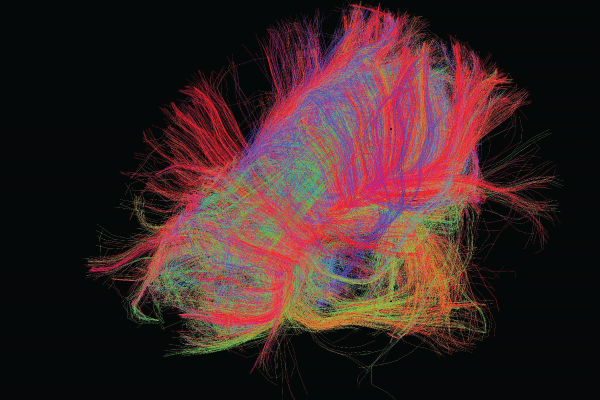
Image courtesy of the USC Laboratory of Neuro Imaging and Athinoula A. Martinos Center for Biomedical Imaging, Consortium of the Human Connectome Project (humanconnectomeproject.org).
Imagine being able to see our brains as a map. By visualizing connections as the equivalents of highways and sideroads serving large cities or rural areas, we could potentially understand how our neural circuits treat information and guide our behavior—as well as how the nervous system’s structure and function are linked. In practical terms, understanding the human brain’s connections and pathways might enable researchers to better understand how certain thoughts and behaviors are originated and maintained. Among other applications, this understanding could provide insights about how the brain develops and ages and how certain diseases or disorders, such as Alzheimer’s disease and psychosis, are related to specific changes in neural pathways.
“The brain is a world consisting of a number of unexplored continents and great stretches of unknown territory.”
Santiago Ramón y Cajal, winner of the 1906 Nobel Prize in Physiology or Medicine
Overview of the Human Connectome Project
The HCP is a massive NIH-funded collaboration to use neuroimaging to map connectivity in the brain. Launched in 2009, it relied on diffusion imaging to trace how various regions of the brain are connected in both healthy individuals and people with various neurological disorders and diseases.
Researchers all over the world continue to use HCP data and the procedural protocols created by the HCP. New computational methods are helping them analyze these data and study them in tandem with other data sets (neuroimaging, genetic, clinical, etc.) to gain new insights on the brain.
Find out more at humanconnectomeproject.org.
There’s only one problem. The creation of such a map—dubbed the “connectome” almost simultaneously by Olaf Sporns of Indiana University and Patric Hagmann of Lausanne University Hospital (Switzerland) in a 2005 instance of “multiple discovery”—would require researchers to map every connection between neurons in the brain and the rest of the human nervous system. That challenge led Sporns and other researchers to start a brand-new field of neuroscience. In an interview with the Observer, Franco Pestilli, a neuroscientist from the University of Texas at Austin, elaborated on the thinking at the time: “The idea of a network—the concept that we shouldn’t be studying only neurons and areas, but also how these areas and these neurons are communicating—resulted in a theoretical shift, a new direction and new needs in neuroscience.”
Although researchers have the technology to completely map the nervous systems of organisms with hundreds to thousands of neurons, mapping the connections among the billions of neurons in the human brain is much more difficult. We know the most about the connectomes of tiny animals, such as worms or larvae. The first such successful mapping was in 1986, when John Graham White, a molecular biologist, along with Sidney Brenner and other colleagues from the University of Cambridge, mapped the 7,000 neuronal connections of the Caenorhabditis elegans, a 1 mm worm that has only 302 neurons. To create the first complete connectome, they used electronic microscopy, deducing the nervous system of the C. elegans from reconstructions of electron micrographs of serial sections.
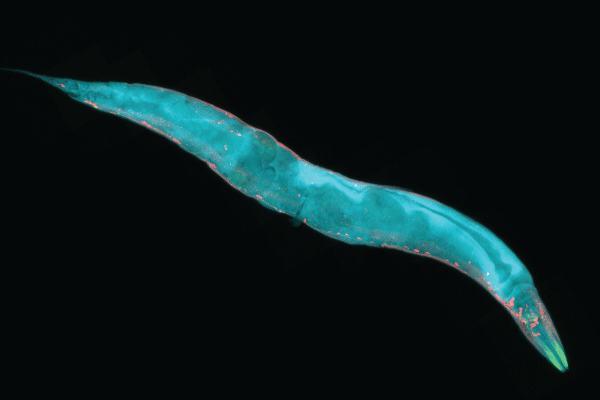
Despite the challenges of mapping the entire human brain network, researchers all over the world are making remarkable progress. One notable effort toward that end is the Human Connectome Project (HCP)—a massive collaboration, funded by the National Institutes of Health (NIH), that seeks to use neuroimaging to map connectivity in the brain.
“The HCP has been a pioneer in the development of algorithms and approaches that map the brain’s connections in exquisite visual detail, allowing us not only to see them but to quantify their differences,” said HCP Co-Principal Investigator Arthur Toga (director of the Mark and Mary Stevens Neuroimaging and Informatics Institute at the University of Southern California) in an email to the Observer. “That’s an important requirement in understanding the differences between populations and individuals.”
The genome on steroids
The human brain contains more than 100 billion neurons (Braitenberg & Atwood, 1958) and trillions of connections. These connections form functional neural networks that underlie all human behavior and cognition.
Pestilli compares the brain to the Internet. Just as billions of computers are connected to the Internet by cables of one kind or another, the billions of neurons in the brain are connected by the “cables” within white matter. The characteristics of those cables define the speed and quality of communication within any brain.
The Connectome Coordination Facility
Funded in 2015, the CCF maintains a central data repository for HCP data and offers advice to the research community. Current CCF studies have three aims:
Studying the healthy young adult brain
These studies support the original data set of 1,200 healthy young adults by distributing image data from multiple imaging modalities, as well as behavioral, demographic, and individual-differences data. The CCF also helps to disseminate HCP-style data acquisition protocols to other studies to maximize comparability among studies.
Studying the brain as it relates to growth and aging
The CCF supports and disseminates protocols for life span studies aimed at mapping how the brain grows and ages. These include studies with babies (ages 0–5), children (ages 5–21), and adults (ages 36–100); studies on the developing prenatal and neonatal brain; and studies on the adolescent brain and cognitive development.
Studying the brain as it relates to disease
These studies apply HCP-style data collection to groups of subjects at risk for or suffering from disorders or diseases affecting the brain. Projects include the study of connectomes relevant to diseases and disorders such as Alzheimer’s disease, aging and dementia, brain degeneration, psychosis, anxiety and depression, visual loss and blindness, and epilepsy, among others.
Find out more about the CCF at humanconnectome.org.
“Fifty percent of the brain volume is neurons, but the other 50% is not—it is the white matter wrapping the neuronal axons—the cables that connect neurons in different areas,” said Pestilli. “But about 80% of neuroscience is about the neurons, and less than 20% of neuroscience is about the cables. The connectome is a lot about these cables and connections.”
From an anatomical point of view, the connectome is “the ensemble over all brain neurons of axonal origin, termination, and trajectory relative to other structures,” wrote Arthur Toga and colleagues in 2012. “Information concerning connectivity is essential for understanding fundamental cognitive operations, systems-level brain activity, conditional structure-function models of brain, and debilitating brain diseases.”
Although “the” human connectome can provide a general map of the human brain, each person’s connectome is unique, even among genetically identical individuals (i.e., identical twins). Moreover, in a 2010 TEDGlobal Talk, the neuroscientist Sebastian Seung (Princeton University) highlighted that every person’s connectome changes over time; structures of neurons change, new synapses are created and others lost, and synapses become larger or smaller. These changes depend, among other factors, on the person’s neural activity (electric and chemical), which in turn depends on their mental experiences, such as perceptions, thoughts, cognitions. Thus, ultimately, a person’s experiences can change their connectome. This means that every connectome is different.
In fact, the term “connectome” was inspired by the efforts to sequence the human genetic sequence—the genome. Just as the genome is mostly the same across all people but shows individual-level variations conferring differences in appearance and health, the human connectome is very similar across people but has individual-level variations in connectivity, reflecting differences in behavior, cognition, personality, and mental health.
One major difference presents an additional challenge to the completion of the human connectome: It’s hundreds of thousands of times larger than the human genome.
What we have mapped
Spanish neuroscientist and artist Santiago Ramón y Cajal (1852–1934) was one of the first researchers interested in mapping the structure of the brain in humans as well as other species, such as birds and cats. At the turn of the 20th century, he developed the first known studies and illustrations of the brain’s microscopic structure. These captured the complex nature of neurons and the central nervous system’s pathways, connections, and functions.
Other advances followed. Ramón y Cajal received the 1906 Nobel Prize in Physiology or Medicine for research in which he subjected thin slices of brain tissue to a silver-nitrate staining procedure (the Golgi method). This allowed him to visualize the nervous system under light microscopy, similar to the method used to capture images on photographic plates. Using a microscope, he saw that neurons resembled “trees growing” and inferred that, given their form, they must conduct electrical information in only one direction—from the dendrites to the axons. He also concluded that each neuron is connected to other neurons and communicates with other neurons through small gaps—the synapses.
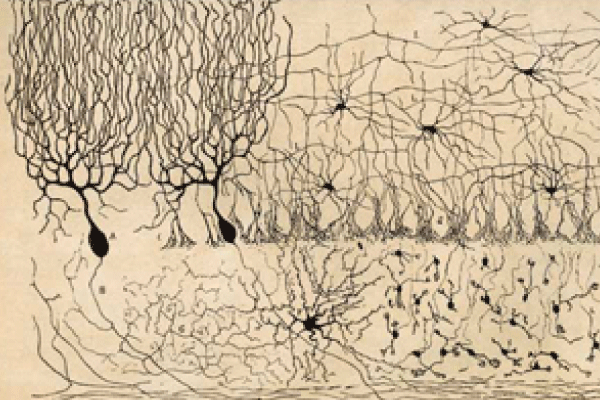
A century later, researchers are still attempting to create a comprehensive map of the complete connectivity structure of the brain. In a 2012 article, Toga and colleagues noted that, for the foreseeable future, a comprehensive description of the complete connectome of even a single human brain might be viewed as unattainable.
In the decades after White and his colleagues mapped the first full connectome of C. elegans in 1986, other researchers computed partial connectomes, including the visual cortex of the macaque (Felleman & Van Essen, 1991), the thalamocortical system in the cat (Scannell et al., 1999), the mouse retina (Briggman et al., 2011) and primary visual cortex (Bock et al., 2011), and the common marmoset’s occipital white matter tracts (Kaneko et al., 2020).
As for mapping the neural pathways of the human brain, it was in 2009 that NIH launched the Human Connectome Project and, within it, two major research consortia to map the human brain’s connections in high resolution, using complementary approaches to map the brain’s wiring. One HCP consortium set out to comprehensively map the brain circuitry of 1,200 healthy adults—twin pairs and their siblings from 300 families—using methods of noninvasive neuroimaging and to provide insights about the organization of brain networks and the genetic and environmental contributions to brain structure and function. The other consortium set out to create a new magnetic resonance scanner optimized for measuring connectome data. This scanner maps the brain’s fibrous long-distance connections by tracking the motion of water, which makes different types of tissues detectable so that the white matter in the brain (where the long extensions of neurons reside) can be sharply seen.
In the intervening years, the HCP has yielded a rich data set on the structural and functional connectivity of a large sample of adults. It has also helped to develop improved methods of imaging, data acquisition, analysis, and sharing. As stated on the NIH website, “HCP has produced stunning maps of neural fibers crisscrossing the brain. It has revolutionized the mapping of connections in the human brain, and has laid foundational groundwork for using brain imaging measures of connectivity as an aid in diagnosis of disease.”
What we can learn—some examples
Using the technical advances provided by the HCP, researchers are expanding their knowledge of the organ’s normal development and aging and assessing disruptions in connectivity that might underlie clinical symptoms. Moreover, they’re able to superimpose demographic, genomic, and cognitive/behavioral data on the connectome to permit inferences about genetic and environmental influences on connectedness.
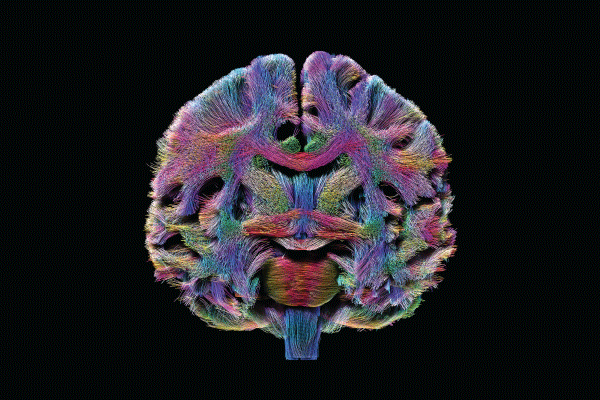
In the area of clinical research, consider the Alzheimer’s Disease Connectome Project. Coordinated by principal investigators Barbara Bendlin (University of Wisconsin–Madison) and Shi-Jiang Li (Medical College of Wisconsin), its goal is to develop robust technology to accurately stage Alzheimer’s disease across its progression in individual subjects. Ultimately, this would contribute to the creation of better tools to evaluate the brain progression of the disease, which affects about 30 million people worldwide and generally progresses for 3 to 9 years after the initial diagnosis, ultimately leading to a complete loss of bodily functioning and death.
Also in the clinical area, the Human Connectome Project for Early Psychosis, coordinated by Alan Breier (Indiana University) and five other researchers, aims to acquire high-quality imaging, behavioral, clinical, cognitive, and genetic data on a group of early psychosis patients and make those data available to the research community for future studies. This could contribute to a better understanding of neural network disruptions in psychotic illnesses and inform more targeted treatment interventions in the illnesses’ early stages, preventing their progression and even chronicity (brain changes are usually not reversible in psychosis). In 2020, the researchers released initial data from the project, including subjects’ structural MRI, resting state fMRI, diffusion MRI, and clinical and behavioral data.
In the developmental area, the Lifespan Human Connectome Project in Development (HCP-D) study aims to advance researchers’ knowledge about how typical development and childhood experiences, such as learning to read or interact socially, can shape brain wiring. This project is coordinated by APS Fellow Deanna Barch (Washington University in St. Louis) and eight other principal investigators. COVID-19 has interrupted data collection, but the researchers are “now starting to look at trajectories of brain development from ages 8 to 21 in terms of cortical thickness and white matter, both by themselves and in relationship to cognitive and affective function,” said Barch in an email to the Observer. The team released their first set of data in 2020, including data from structural MRI, resting-state fMRI, task fMRI, and diffusion MRI for more than 800 subjects.
HCP protocols can also support research aimed at developing understanding of cognitive processes that may be impaired in clinical populations. In a project called Dual Mechanisms of Cognitive Control, a team led by APS Fellow Todd Braver (Washington University in St. Louis) focuses on understanding the psychological and neural mechanisms that give rise to cognitive-control processes, which are not only important for domains such as attention, working memory, episodic memory, and decision-making but are also thought to be sources of functional impairment for individuals who suffer from mental or neuropsychiatric disorders. This project is relevant to public health because it can provide information on the brain basis of normal human variation in mental functions such as attention, memory, decision-making, and intelligence. This could improve understanding of the relationship between normal functioning and mental health disorders and the risk factors for such disorders.
Studying brain connections can also tell us which connections are “good” or “bad.” Pestilli and his lab have been applying new technologies to understand white matter and advance the neuroanatomical understanding of the brain. White matter—oligodendrocytes—is the matter that wraps axonal projections in the brain, influencing the axons’ quality, explained Pestilli. Going back to his Internet metaphor, just as cables can be made of different materials ranging from copper (slow and unreliable) to optical fiber (fast and reliable), white matter can have different structures that make connections between neurons fast or slow, reliable or unreliable.
Pestilli’s lab has done computational work to analyze these connections and distinguish “good” and “bad” connections. This type of analysis can also bring clarity to neuroanatomy. For instance, Pestilli and his colleagues identified a connection between the ventral and dorsal streams in the brain (supposedly responsible for “what” and “where” information, respectively, in the visual system). This connection—the vertical occipital fasciculus—is located early in the streams, suggesting that the two streams are highly interactive very early on. Curiously, other researchers suggested this connection many years ago, but it took new technologies to actually prove it. This finding clarifies a structural aspect of the brain, with implications for cognition. Pestilli’s lab is currently exploring whether human cognition might have a basis in dorsal-ventral communication.
The future of network neuroscience
“The connectome is perhaps one of the most groundbreaking theories, changes, revolutions in the last 20 to 30 years of neuroscience,” said Pestilli. A groundbreaking aspect of the HCP, in turn, is that it has advanced the field at the level of informatics and through culture changes such as the push for data sharing, he believes.
Other major shifts have occurred in that time. For example, the study of networks in the brain has expanded, and today the rapidly growing field of network neuroscience is “attracting physicists, highly computationally skilled trainees, and experts in the field of neuroscience,” Pestilli said. There is also the ongoing push to improve reproducibility by allowing the reuse of data, code, and informatics tools. He credits APS Fellow Russ Poldrack (Stanford University) as a major driver behind this push, noting that the goal is to not only “promote reproducibility but also accelerate discovery by allowing easy reuse of assets.”
Perhaps not surprisingly, the combination of a fast-growing interdisciplinary field and the push to make data and tools more shareable and reusable has made neuroscience more complicated. “The complexity of analysis is growing day by day,” Pestilli said.
Feedback on this article? Email apsobserver@psychologicalscience.org or scroll down to comment.
Published in the print edition of the March/April issue with the headline “Totally Wired.”
References
Bock, D. D., Lee, W.-C. A., Kerlin, A. M., Andermann, M. L., Hood, G., Wetzel, A. W., Yurgenson, S., Soucy, E. R., Kim, H. S., & Reid, R. C. (2011). Network anatomy and in vivo physiology of visual cortical neurons. Nature, 471(7337), 177–182.
Braitenberg, V., & Atwood, R. P. (1958). Morphological observations on the cerebellar cortex. Journal of Comparative Neurology, 109(1), 1–33.
Briggman, K. L., Helmstaedter, M., & Denk, W. (2011). Wiring specificity in the direction-selectivity circuit of the retina. Nature, 471(7337), 183–188.
Felleman, D. J., & Van Essen, D. C. (1991). Distributed hierarchical processing in the primate cerebral cortex. Cerebral Cortex, 1(1), 1–47.
Hagmann, P. (2005). From diffusion MRI to brain connectomics (Thesis No. 3230)[Doctoral dissertation, École Polytechnique Fédérale de Lausanne]. EPFL. https://doi.org/10.5075/epfl-thesis-3230
Kaneko, T., Takemura, H., Pestilli, F., Silva, A. C., Frank, Q. Y., & Leopold, D. A. (2020). Spatial organization of occipital white matter tracts in the common marmoset. Brain Structure and Function, 225(4), 1313–1326.
Scannell, J. W., Burns, G. A. P. C., Hilgetag, C. C., O’Neil, M. A., & Young, M. P. (1999). The connectional organization of the cortico-thalamic system of the cat. Cerebral Cortex, 9(3), 277–299.
Seung, S. (2010, July). I am my connectome [Video]. TED Conferences. https://www.ted.com/talks/sebastian_seung_i_am_my_connectome
Sporns, O., Tononi, G., & Kötter, R. (2005). The human connectome: A structural description of the human brain. PLoS Computational Biology, 1(4), Article e42.
Toga, A. W., Clark, K. A., Thompson, P. M., Shattuck, D. W., & Van Horn, J. D. (2012). Mapping the human connectome. Neurosurgery, 71(1), 1–5.
White, J. G., Southgate, E., Thomson, J. N., & Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society B: Biological Sciences, 314(1165), 1–340.
Project Closeup: The Lifespan Human Connectome Project in Development (HCP-D)
Coordinated by APS Fellow Deanna Barch (Washington University in St. Louis), HCP-D leverages technological and analytical progress in brain imaging to chart developmental changes in brain connectivity at unprecedented levels of detail. A longitudinal component focuses on the pubertal period (ages 9–17).
Project overview
• Participants are approximately 1,400 healthy 5- to 21-year-olds in four sites around the United States from diverse geographical, ethnic, and socioeconomic backgrounds.
• Brain imaging sessions are acquired using a 3T Siemens Prisma platform and include structural, functional (resting-state and task-based), diffusion, and perfusion imaging.
• Behavioral measures are a battery of cognitive tasks and self-reports. The parents of minor participants also complete a battery of instruments to characterize cognitive and emotional development and environmental variables relevant to development.
• Participants also provide biological samples of blood, saliva, and hair, enabling assays of pubertal hormones, health markers, and banked DNA samples.
• HCP-D will ultimately comprise approximately 1,750 open-access data sets.
Building on the HCP
“We used a range of imaging approaches, including measures of gray matter structure, task-based imaging (measures of emotion, cognitive control, and reward processing), and both structural and functional connectivity,” Barch explained. “These methods all built upon the advances of the first Human Connectome Project, but with adaptations for development, including an even stronger focus on controlling movement and being able to address movement.”
“There are many projects now looking at brain connectivity in a variety of conditions across the life space (depression, anxiety, psychosis, etc.), and our data help inform typical developmental patterns to which data in these other populations can be compared,” she said.
The next challenge Barch defines the next big challenge for HCP-D as developing “methods to harmonize these data with the data from the original young adult Human Connectome Project, since the scanners changed and the initial HCP used slightly different protocols. In general, this will be the challenge for all of the [HCP] projects—how to integrate the data across different populations and platforms to develop a really integrated picture of brain development across the life span.”
“We are going to focus on sophisticated analyses of the data, particularly the relationships to puberty, which is really critical to understanding both typical development and conditions that tend to arise as puberty evolves, such as depression or suicidal ideation,” Barch said.
Wiring in action: Mapping the impact of collision sports
Athletes in many collision sports, especially American football, are exposed to repetitive head impacts that may affect the structure of the brain even if they don’t result in diagnosed concussions. Franco Pestilli’s lab has performed several studies on these impacts, including investigating whether college football players, cross-country runners, and non-athlete students show differences in their brains’ white matter structures, which determine the quality and type of connections between neurons.
In one study, the researchers used an anatomically informed, personalized-medicine tractography approach to determine which major white matter tracts showed the greatest degree of difference in tensor measures among these three student populations (Caron et al., 2018). Results indicated that differences in white matter, especially in longer fiber tracts, were greatest for football players. These findings suggest that head impacts’ effects may depend on the geometric properties of white matter tracts and support the hypothesis that multiple head impacts can cause structural changes in white matter that are detectable with diffusion MRI and tractography.
In another study—the first investigation into the effects of repetitive head impacts using open-source data-processing platform Brainlife.io—researchers in Pestilli’s lab used two models of the diffusionweighted MRI signal (Caron et al., 2020). Each model’s parameters were mapped in both cortical and subcortical brain structures, as well as in the major white matter tracts. Both models showed that football players had consistently higher measures of microstructure than non-athletes, possibly because of neuroinflammatory mechanisms.
References
Caron, B., Bullock, D., Kitchell, L., McPherson, B. C., Kellar, D. A., Cheng, H., Newman, S., Port, N., & Pestilli, F. (2020). Advanced mapping of the human white matter microstructure better separates elite sports participation. PsyArXiv. https://doi.org/10.31234/osf.io/dxaqp
Caron, B., Port, N., & Pestilli, F. (2018). Advanced white matter mapping in the subconcussive brain. Neurology, 91(23 Supplement 1), S15–S15.





APS regularly opens certain online articles for discussion on our website. Effective February 2021, you must be a logged-in APS member to post comments. By posting a comment, you agree to our Community Guidelines and the display of your profile information, including your name and affiliation. Any opinions, findings, conclusions, or recommendations present in article comments are those of the writers and do not necessarily reflect the views of APS or the article’s author. For more information, please see our Community Guidelines.
Please login with your APS account to comment.